 PubMed Central (PMC) was established in 2000 as the National Library of Medicine’s full-text, journal article repository. Since 2005, PMC has also been the designated repository for papers submitted in accordance with the NIH Public Access Policy. Today, PMC serves as the full-text repository for papers across a variety of scientific disciplines that fall under a number of funding agencies’ public access policies.
PubMed Central (PMC) was established in 2000 as the National Library of Medicine’s full-text, journal article repository. Since 2005, PMC has also been the designated repository for papers submitted in accordance with the NIH Public Access Policy. Today, PMC serves as the full-text repository for papers across a variety of scientific disciplines that fall under a number of funding agencies’ public access policies.
These public access policies seek to make the published full-text papers, resulting from publicly- and privately-funded research, available for the public to find and read. As a repository, PMC ensures the permanent preservation of these research findings and makes the results of this research more readily accessible to the public, healthcare providers, educators, and the scientific community.
Recently PMC updated its policy statement on supplementary data to more clearly articulate the requirement that any supplementary data (images, tables, video, or other documents/files) that are associated with an article must be deposited in PMC with an article.
This applies to all files made available in the article record, even if the files are also available in a public repository. An exception may be made for data files that require custom software to read and use, or are very large (over 2 GB).
In cases where data cannot be reasonably included with an article, either in a figure, table, or supplementary file, NLM encourages journals and authors to make the data available in a public repository and include the relevant data citation(s) in the paper.
The NIH Manuscript Submission (NIHMS) system, developed to facilitate the submission of peer-reviewed manuscripts for inclusion in PMC, can accept submissions of datasets (2 GB or smaller) in support of any manuscript files deposited in compliance with a participating funder’s public access policy. Because these datasets will be publicly accessible, those related to human subjects research should not include any personally identifiable information and deposit should be consistent with informed consent. For more information on depositing supplementary data and dataset files via NIHMS, see the related NIHMS FAQ.
For questions regarding this revised policy or for guidance with depositing supplementary data, please refer to the HSLS Scholarly Communication: Public Access Policies page or contact HSLS Data Services.
*Parts of this article were derived from PMC documentation: Funders and PMC and PMC Policies
~Melissa Ratajeski
 The week of February 11–15, 2019, is Love Data Week. To celebrate, HSLS Data Services will be hosting a variety of activities, workshops, and giveaways throughout the week. Stop by the library to engage with colleagues and librarians and learn best practices and tips for loving your data.
The week of February 11–15, 2019, is Love Data Week. To celebrate, HSLS Data Services will be hosting a variety of activities, workshops, and giveaways throughout the week. Stop by the library to engage with colleagues and librarians and learn best practices and tips for loving your data.
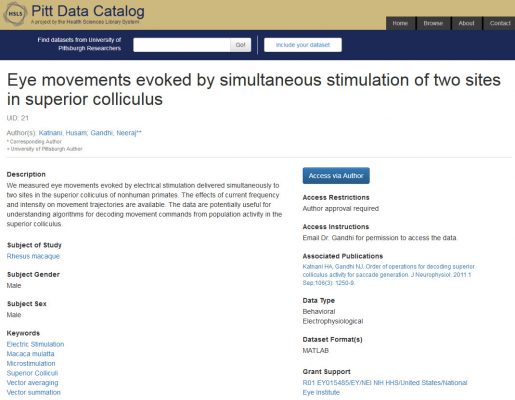




 As
As  The groups worked for three long, collaborative, and productive days, capped with irreverent awards such as “best hair” and “how I learned to relax and love the hackathon” (see picture). Final projects included:
The groups worked for three long, collaborative, and productive days, capped with irreverent awards such as “best hair” and “how I learned to relax and love the hackathon” (see picture). Final projects included: