 There are challenges with downloading genomic data. File sizes are large, and it can be time consuming to retrieve multiple files. Sometimes downloads fail. A custom script may be required. Fortunately, a solution to all of these frustrations is now available—NCBI Datasets.
There are challenges with downloading genomic data. File sizes are large, and it can be time consuming to retrieve multiple files. Sometimes downloads fail. A custom script may be required. Fortunately, a solution to all of these frustrations is now available—NCBI Datasets.
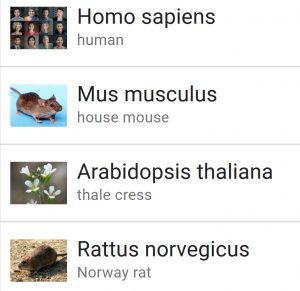
This experimental resource allows users to easily download eukaryotic genome sequence and annotation data by assembly accession, taxonomic name (scientific and common), or taxonomy ID. The web interface allows for browsing by organism, with the most common experimental species conveniently available from the main page. For example, try selecting the house mouse (mus musculus), then select all 22 associated assemblies. Options for the type of data for the download include genomic, transcript, and protein sequences as well as annotation features.
When a selected dataset is close to or above the limit of 15 GB, the downloaded file will be a “dehydrated bag,” aka a compressed/zipped file containing only the data report and links to download the selected dataset(s) from the NCBI servers.
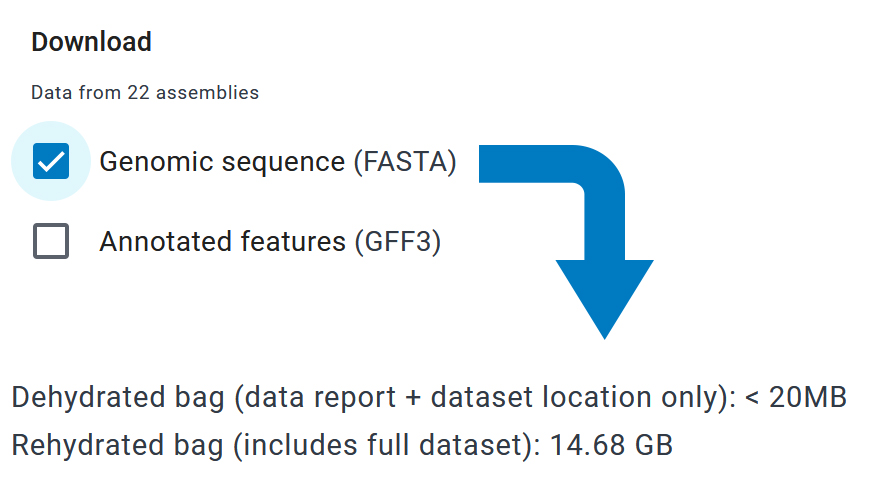
Instructions are provided to “rehydrate” the unzipped files and access the full dataset(s). This dehydrate/rehydrate strategy makes it simple to download, share, and store large genome datasets. For example, sharing this data with a colleague is as easy as e-mailing the dehydrated file, which can then be rehydrated at a convenient time.
Plans for NCBI Datasets include adding other assemblies (bacteria and viruses) and datasets such as genomic patch sequence and alternative loci. The project aims to meet the FAIR principles of scientific data management (Findable, Accessible, Interoperable, and Reusable). An introduction to NCBI Labs initiatives is available on the NCBI Insights blog.
Use NCBI Datasets to gather genomic data in order to practice using the many bioinformatics tools licensed by HSLS. Need help with using these resources? Contact HSLS MolBio or (virtually) attend one of our many workshops.
~Carrie Iwema