 As previously reported, HSLS hosted a National Center for Biotechnology Information (NCBI) Hackathon from September 25-27, 2017, in collaboration with numerous campus partners. The event took place in the Digital Scholarship Commons of the University Library System (ULS). HSLS, the Center for Research Computing (CRC), and the Department of Biomedical Informatics (DBMI) generously provided support for breakfasts. Computing Services and Systems Development (CSSD), the School of Computing and Information (SCI), and the CRC provided expert technical support.
As previously reported, HSLS hosted a National Center for Biotechnology Information (NCBI) Hackathon from September 25-27, 2017, in collaboration with numerous campus partners. The event took place in the Digital Scholarship Commons of the University Library System (ULS). HSLS, the Center for Research Computing (CRC), and the Department of Biomedical Informatics (DBMI) generously provided support for breakfasts. Computing Services and Systems Development (CSSD), the School of Computing and Information (SCI), and the CRC provided expert technical support.
An NCBI-style Hackathon is a social event in which highly motivated individuals with expertise in scientific disciplines, computer programming, software development, etc., meet for an intense few days to formulate useful, efficient pipelines supporting biomedical research. All code generated by NCBI-Hackathons is made freely available on GitHub, and manuscripts describing the design/usage of software tools are posted on the F1000Research Hackathons channel.
The Pitt/NCBI-Hackathon was led by Ben Busby, the NCBI Genomics Outreach Coordinator. Participants were primarily from Pittsburgh, but they also traveled from Columbus, Oh.; Baltimore, Md.; Charlottesville, Va.; New York, N.Y.; Denver, Colo.; and San Diego, Calif. Initially, the 24 hackers were divided into five teams, but two of the groups working on virus discovery and identification of past viral exposure merged to form a super-group—an NCBI-Hackathon first!
 The groups worked for three long, collaborative, and productive days, capped with irreverent awards such as “best hair” and “how I learned to relax and love the hackathon” (see picture). Final projects included:
The groups worked for three long, collaborative, and productive days, capped with irreverent awards such as “best hair” and “how I learned to relax and love the hackathon” (see picture). Final projects included:
- HAQmap—a guide containing information and tools to help organizers create their own NCBI-style hackathon (5 member team).
- (SC)3 Super Concise Single Cell SNP Caller—this project enables finding expressed SNPs in SRA data associated with a Bioproject record (3 member team).
- SPeW: SeqPipeWrap—a framework for taking a NextGen Seq pipeline (such as RNA-seq, ChIP-seq or ATAC-seq) in any language, and using NextFlow as a pipeline management system to create a flexible, user-friendly pipeline that can be shared in a container platform (6 member team).
- ViruSpy—a pipeline designed for virus discovery from metagenomics sequencing data available in NCBI’s SRA database (10 member team).
The success of the Pitt/NCBI-Hackathon bodes well for the possibility of future hackathons. If you are interested in learning more, please contact the HSLS Molecular Biology Information Service.
~ Carrie Iwema

 Users of PubMed and other databases provided by the National Center for Biotechnology Information (NCBI) can save searches, citations, interface preferences, and other information in a MyNCBI account. When MyNCBI debuted there was only one option, to create a new username and password from scratch. NCBI calls this a “native account.” Now MyNCBI is connected to other NIH systems such as the eRA Commons and the SciENcv research biosketch service, and the options for logging into MyNCBI have expanded to include 3rd party logins tied to the user’s Google, eRA Commons, ORCID, University of Pittsburgh, or other accounts. Is there an advantage to creating a new account in MyNCBI? Why not use the University of Pittsburgh 3rd party login and eliminate one more password you must remember?
Users of PubMed and other databases provided by the National Center for Biotechnology Information (NCBI) can save searches, citations, interface preferences, and other information in a MyNCBI account. When MyNCBI debuted there was only one option, to create a new username and password from scratch. NCBI calls this a “native account.” Now MyNCBI is connected to other NIH systems such as the eRA Commons and the SciENcv research biosketch service, and the options for logging into MyNCBI have expanded to include 3rd party logins tied to the user’s Google, eRA Commons, ORCID, University of Pittsburgh, or other accounts. Is there an advantage to creating a new account in MyNCBI? Why not use the University of Pittsburgh 3rd party login and eliminate one more password you must remember?
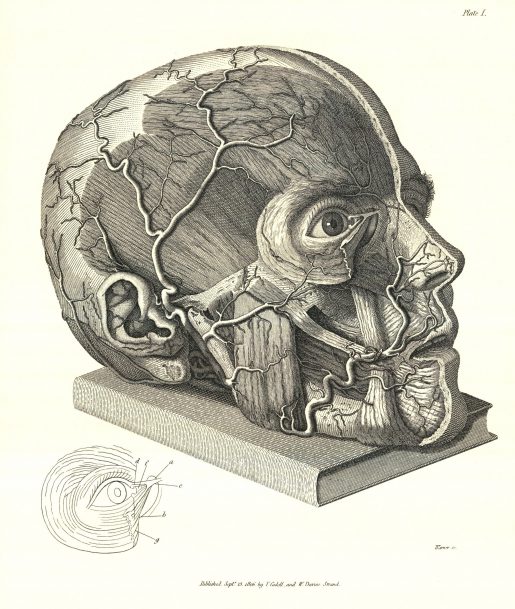 Although he was not a loveable person, his innovative teaching methods, and the excellent books he authored, earned him the respect of the medical profession. He is remembered as a great surgeon, anatomist, and discoverer of the naso-palatine nerve. His study of the hearing and olfactory organs is considered classic. He was the first to correctly delineate the nerves of the heart. In his authoritative works on hernia, he described perineal hernia from direct observations, thus settling any earlier controversy of its existence. His work, Saggio di osservazioni e d’esperienze sulle principali malattie degli occhi (Practical observations on the principal diseases of the eyes, 1806), raised ophthalmology to the level of an autonomous science. Plate I from this work is one of the most famous illustrations in medicine, in which science and art become one.
Although he was not a loveable person, his innovative teaching methods, and the excellent books he authored, earned him the respect of the medical profession. He is remembered as a great surgeon, anatomist, and discoverer of the naso-palatine nerve. His study of the hearing and olfactory organs is considered classic. He was the first to correctly delineate the nerves of the heart. In his authoritative works on hernia, he described perineal hernia from direct observations, thus settling any earlier controversy of its existence. His work, Saggio di osservazioni e d’esperienze sulle principali malattie degli occhi (Practical observations on the principal diseases of the eyes, 1806), raised ophthalmology to the level of an autonomous science. Plate I from this work is one of the most famous illustrations in medicine, in which science and art become one. A new website called,
A new website called, 