Are you a biomedical researcher searching for information on genes related to a particular disease, pathway, or process? Are you overwhelmed by the vast quantity of associated literature? Have you considered that some genes are more highly investigated than others, which leads to an overabundance of literature to review for some genes as well as a scarcity of information on others?
Geneshot is a new search engine created to bridge this gap and highlight understudied genes by mining publications for mentions of any genes with the search term(s), then prioritizing the genes. Searching for a biomedical term returns two ranked gene lists. The first contains genes reported in the literature and the second contains predicted genes identified via data integration from multiple sources. The underlying data sets are from PubMed, GeneRIF, AutoRIF (more comprehensive than the former), gene-gene co-expression matrix data using ARCHS4, and gene-gene co-occurrence matrix Tagger and Enrichr data. Geneshot can also facilitate hypothesis generation by assessing gene set novelty and proposing additional relevant genes to augment gene sets. A detailed description of the mining and prediction methodology is available in the Geneshot publication.
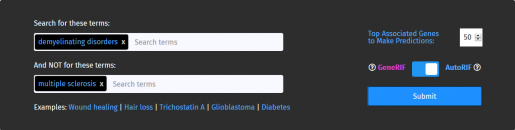
To use Geneshot:
- Enter the search term(s) (e.g., demyelinating disorders) as well as any terms not to be included in the search (e.g., multiple sclerosis).
- Select the number of top associated genes to generate the list of predicted genes; default is 50.
- Select searching by GeneRIF or AutoRIF.
Click the Submit button.
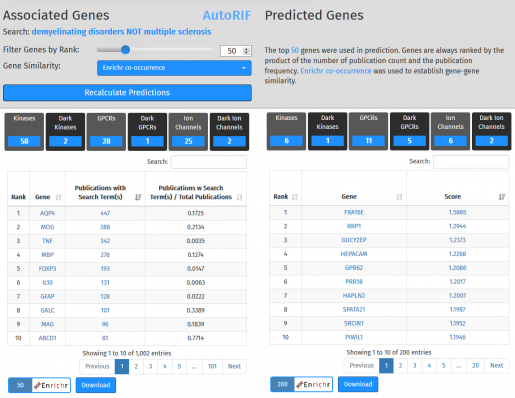
Search results include:
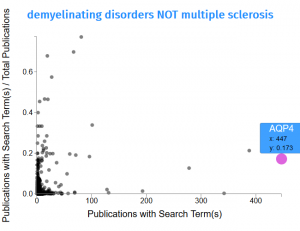
- A scatterplot showing the genes associated with the search term. Each dot represents a gene. In this example, the gene AQP4 is associated with 447 publications that also match the search terms “demyelinating disorders NOT multiple sclerosis” (x: 447). From all publications that mention AQP4, 17.3% also include the search parameters.
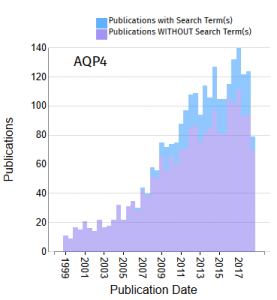
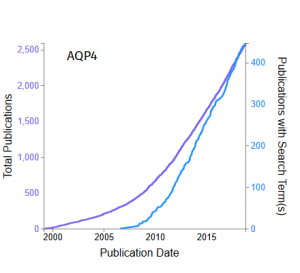
- A histogram or cumulative distribution plot of the association of a selected gene with the search term over time.
- Two modifiable gene tables containing the Associated or Predicted gene lists as described previously. Clicking on a gene name opens a page with additional information, including functional associations. Clicking on a publications number opens a PubMed results page. Clicking on a score shows the top ten genes that caused that gene to be predicted to be associated with the search term.
Geneshot was created by the Ma’ayan Laboratory at the Icahn School of Medicine at Mount Sinai.
~Carrie Iwema