iCite, from the National Institutes of Health (NIH), is a powerful web-based tool that provides bibliometric information for journal articles included in PubMed. Users can analyze single or multiple articles via three modules:
1. Influence:

Delivers scientific influence metrics using the Relative Citation Ratio (RCR). Developed by the NIH, the RCR is a metric that represents the citations/year of a paper, normalized to the citations/year received by NIH-funded papers in the same field and year. For example, a paper with an RCR of 1.0 has the same number of citations/year as the median NIH-funded paper in its field, whereas a paper with an RCR of 2.0 has twice as many citations/year. For more information about how RCR is calculated, read these two articles by B. Ian Hutchins, “Relative Citation Ratio (RCR): A New Metric That Uses Citation Rates to Measure Influence at the Article Level” (2016) and “Additional Support for RCR: A Validated Article-level Measure of Scientific Influence” (2017).
2. Translation:
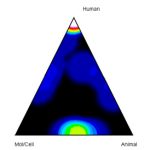
Measures a paper’s research subject orientation (human, animal, or molecular/cellular biology) to track and predict future citations by clinical articles using the Approximate Potential to Translate (APT), which is a machine learning-based estimate of the likelihood that a paper will later be cited in clinical trials/guidelines. Read more about APT and predicting translational progress in biomedical research in B. Ian Hutchins’ article, “Predicting Translational Progress in Biomedical Research” (2019).
3. Open Cites:
Disseminates link-level, public-domain citation data from the NIH Open Citation Collection (NIH-OCC). The NIH-OCC is a public access database for biomedical research aggregating open citation data gathered from MEDLINE, PubMed Central, and CrossRef. Read more about the NIH-OCC in B. Ian Hutchins’ article, “The NIH Open Citation Collection: A Public Access, Broad Coverage Resource” (2019).
All three modules are analyzed simultaneously with a given search, with multiple options for the search query: (1) Search PubMed by author name, title, MeSH keyword, etc., (2) Upload a spreadsheet of PubMed Identifiers (PMIDs), or (3) Input a list of PMIDs. A maximum of 10,000 PMIDs may be queried at a time, and the iCite database coverage is limited to 1980-current.
Results are displayed in a tabbed format by module, with customization options that include limiting by publication year(s) and/or by article type (research only, clinical only, and/or cited by clinical articles only). All relevant articles (or the most recent 10,000) are also listed, including PMID links and RCR scores. Searches may be re-analyzed using the options “Citing Papers” (run a new analysis on the papers that cite these articles) and “Referenced Papers” (run a new analysis on the papers that are referenced by these articles).
~ Carrie Iwema