Cufflinks & Cuffmerge & Cuffdiff, oh my! (content from Sequencher)
One of the hottest topics in laboratory science is the rapidly expanding field of RNA-Seq. Fueled by ever more efficient NGS instruments now available to researchers, it is possible to get a much more accurate snapshot of a cell’s transcriptome by isolating and sequencing RNA. Even without the complexity of gene regulation, the presence of splice variants and the technical issues involved in dealing with RNA, characterizing the transcriptome using command line tools can be a daunting task. The Cufflinks(1) suite of programs, which is one of the most peer reviewed RNA-Seq tools for examining differential gene expression, has now been added to Sequencher.
Cufflinks not only finds the relative abundance of mRNA transcripts, but also looks for different isoforms of the same gene which can give insight into complex gene regulation through splicing and other forms of RNA editing. Researchers at ITQB(2) used Cufflinks to examine how the steady state levels of RNA in E.coli are affected by three common exoribonucleases (RNase II, RNase R, and PNPase). Comparing the gene expression data from cultures that each had knock downs for one of the exoribonucleases the researchers were able to find correlations between the nucleases and which transcripts they could bind to.
The researchers were also able to tie the downstream results of the gene expression knock down to several important features. The features noticeably affected were bacterial mobility and biofilm formation which are both important in factors in bacterial pathogenicity.
Cufflinks can be used to find non-coding RNAs as well. An article from the European Heart Journal3 cited research that identified hundreds of novel long non-coding RNAs that played specific roles in cardiac function after a heart attack. Finding the relationship between the lncRNA and the gene regulation they are involved in could lead to a breakthrough in new medical treatments for heart attack victims.
If you are starting with raw cDNA data, you can perform every step of your RNA-Seq analysis with Sequencher. Your first step will be choosing which NGS alignment algorithm to use. Sequencher has integrated BWA-MEM and GSNAP with easy to use graphic interfaces. You will still have access to all the command line arguments just as you would have if running them from the command line.
The NGS alignment will produce a SAM/BAM file which is the input file to the Cufflinks workflow. The Cufflinks workflow has three steps. Normally these steps are performed on the command line where case-sensitivity, mistaking tabs for white space (or vice versa), abbreviations or single letters for options, leave no room for error. Sequencher removes the command line requirement and gives the user an easy to use graphical interface. The Cufflinks program is the first step which takes the aligned reads from the SAM file and re-aligns them to the gene model in the GTF annotation file. It also looks for different isoforms as well as novel transcripts. The abundance of the transcripts is related to the normalized number of reads. If you are looking at differential expression then this step is repeated for each of the two samples or condition that is being compared.
The next step is called Cuffmerge, and like the name implies this will merge the two transcript files generated from Cufflinks into a single merged consensus file of transcripts. This file will be used in the final step with Cuffdiff which performs the differential expression analysis.
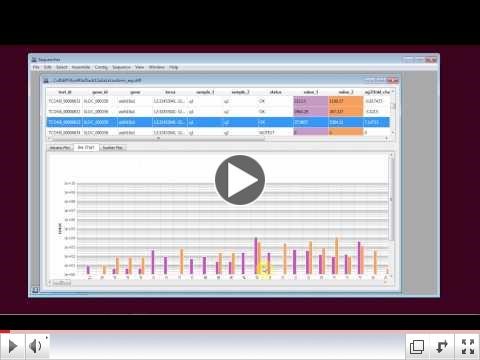
Sequencher takes the output files from Cuffdiff and generates a number of graphical views to illustrate the differences in the expression levels. This is another area where Sequencher saves the user from the command line as well as the statistical programming language R. The three graphical views that Sequencher provides are a volcano plot, scatter plot, and a bar chart. The volcano plot can be especially useful in determining points of interest as it plots the magnitude of the change in expression versus the statistical relevance of that change. Each graph highlights a different aspect of the analysis and all the graphs are linked to the data displayed in the table above it. If you click on a point on a graph, it will automatically highlight which data entry is responsible for that point.
The addition of Cufflinks is just another example of Gene Code’s commitment to deliver features that our customers request. Gene Codes has greatly expanded the platform of tools that Sequencher offers and will continue to do so going forward. The continued expansion of NGS and RNA-Seq functionality will further enhance the value and performance of Sequencher software.
Citations